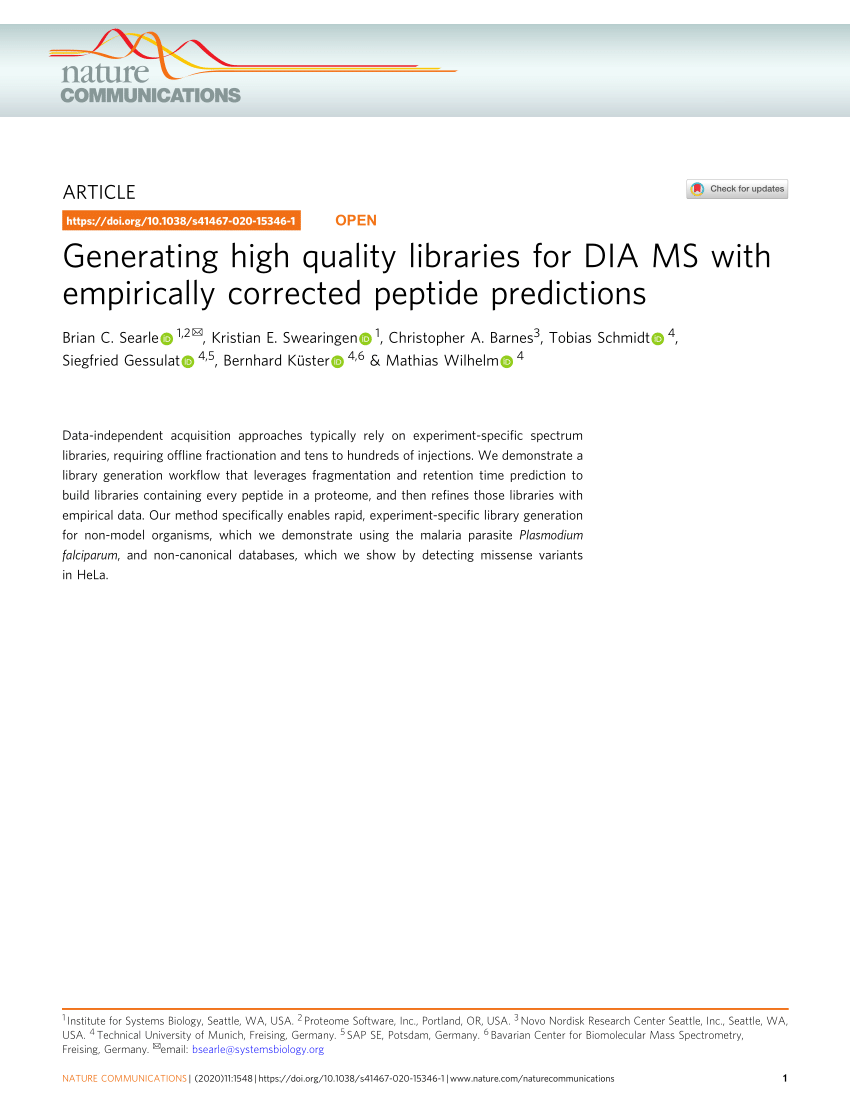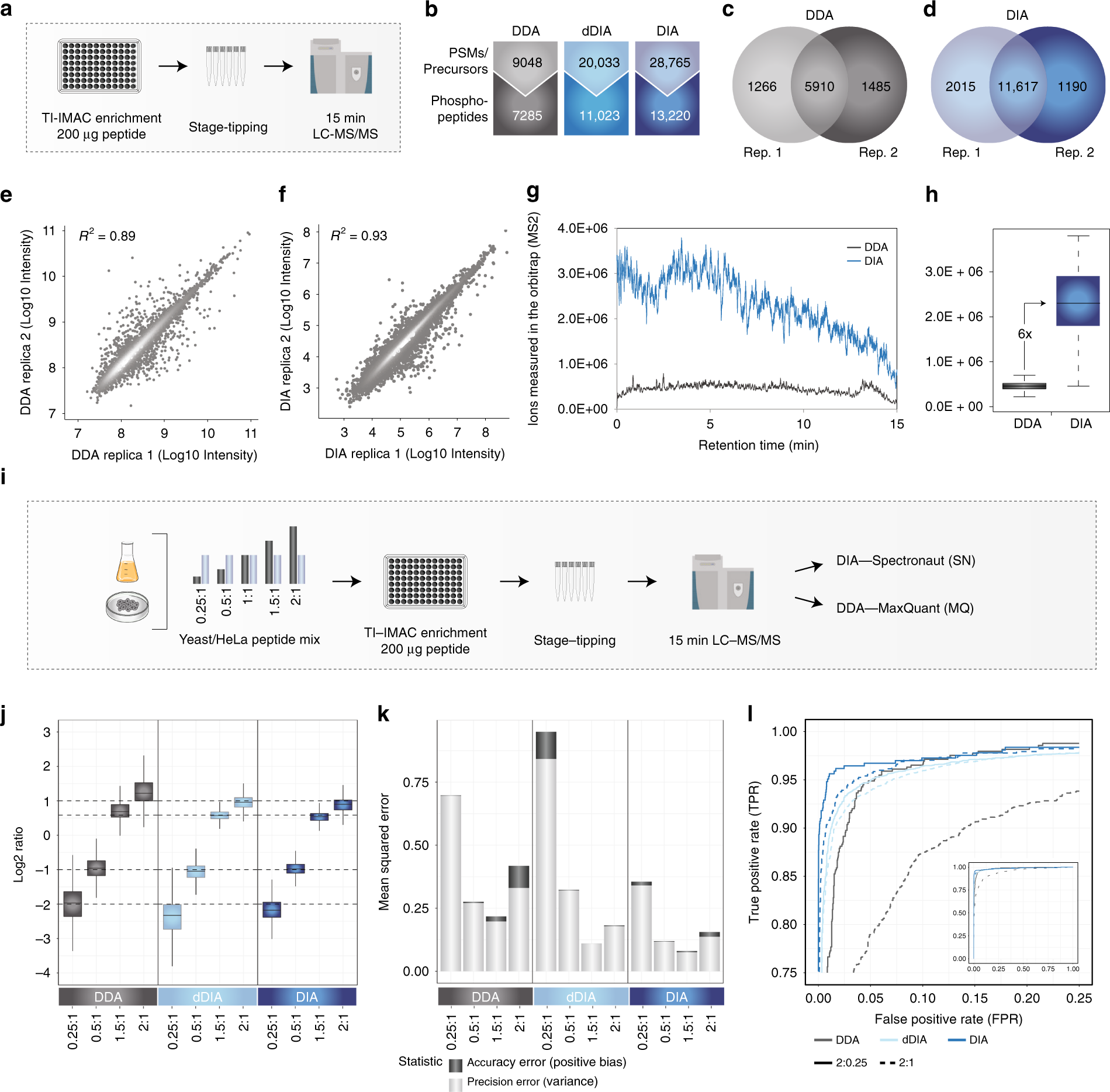
Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries | Nature Communications
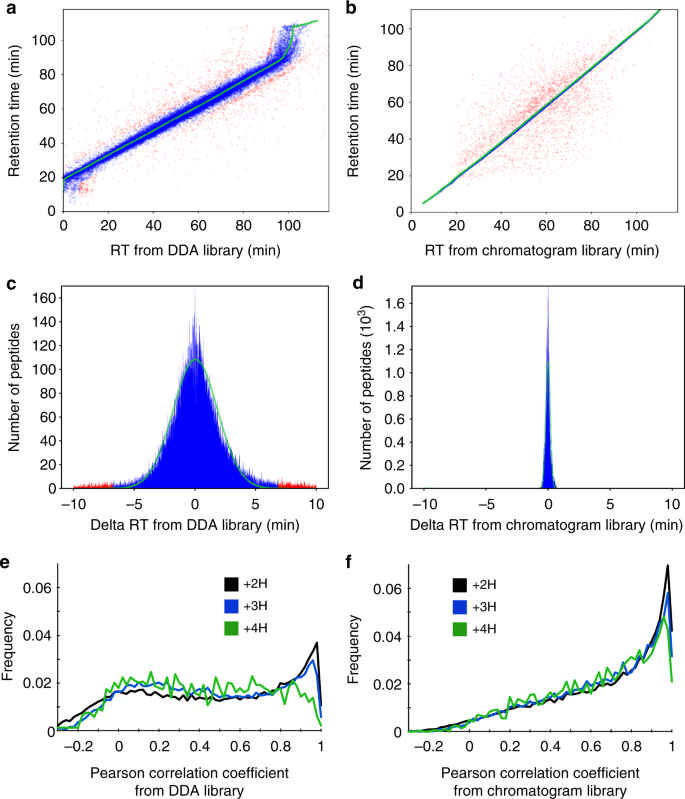
Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry | Nature Communications
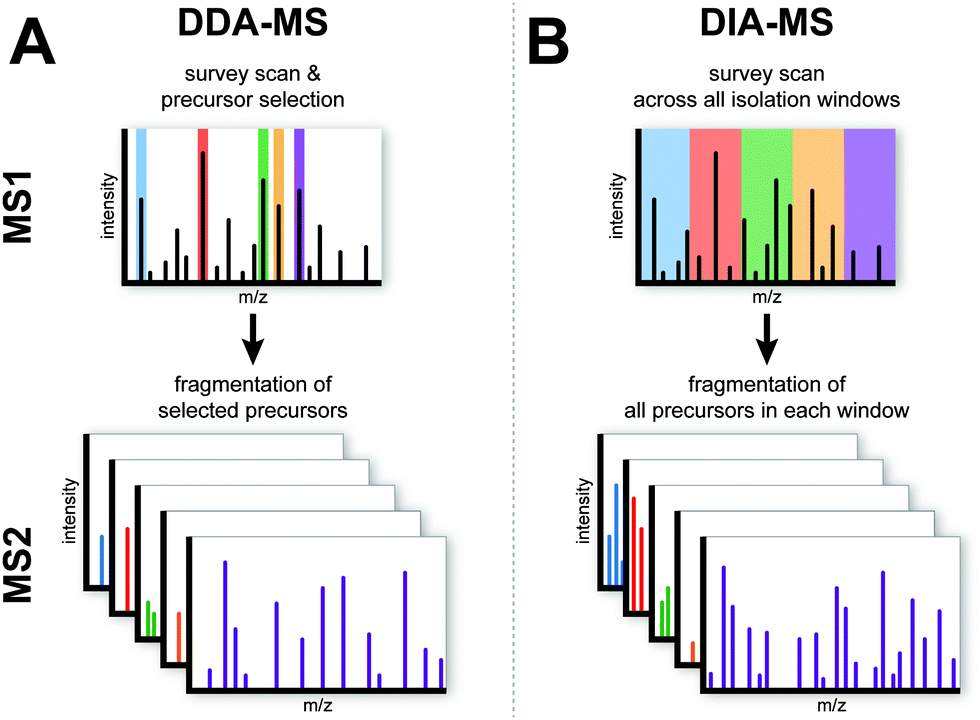
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H
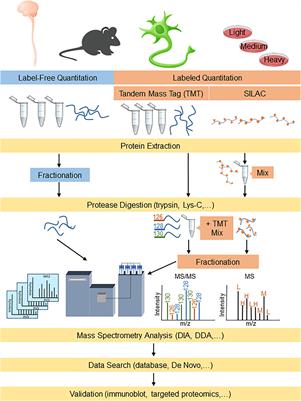
Frontiers | Proteomics Approaches for Biomarker and Drug Target Discovery in ALS and FTD | Neuroscience
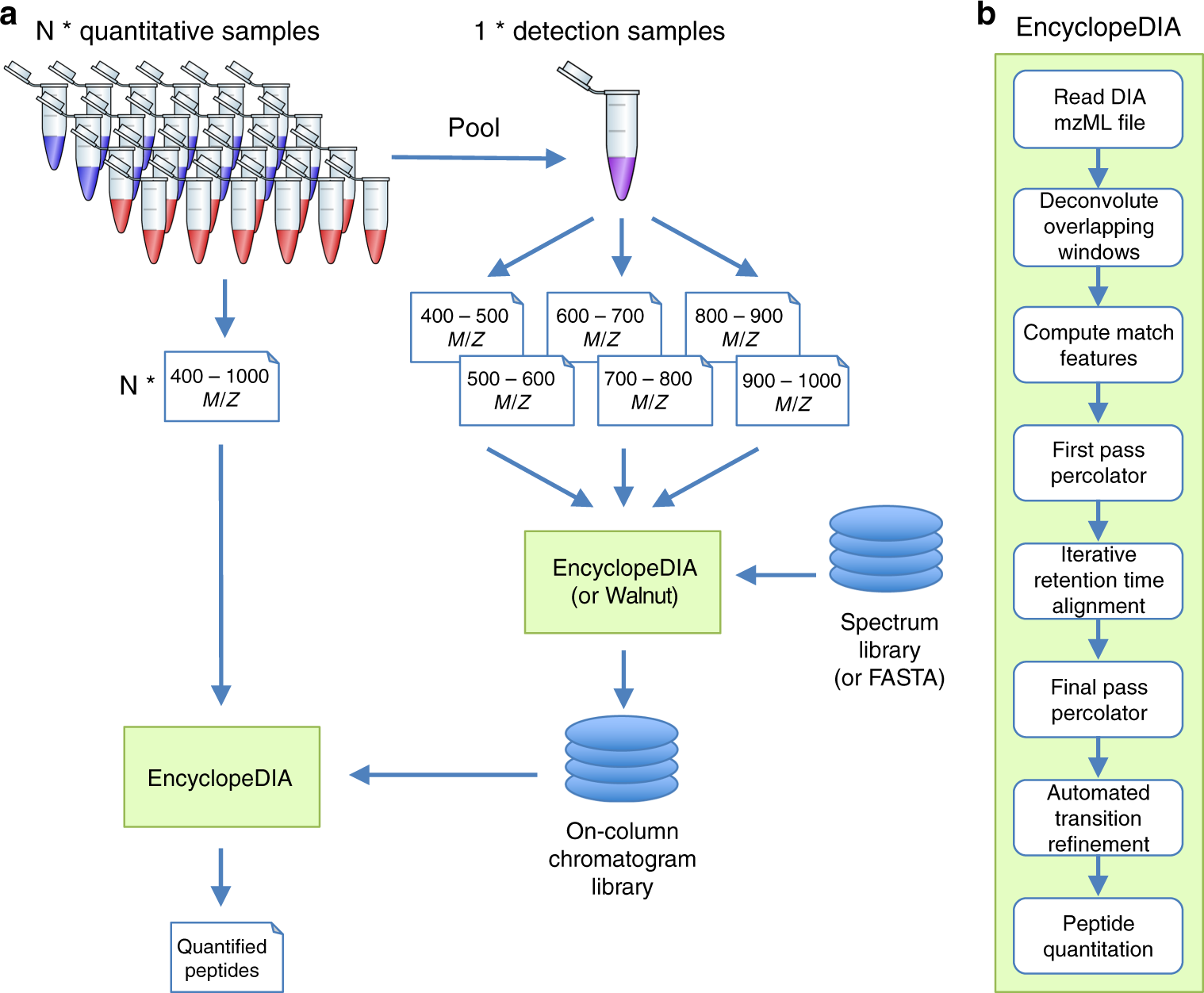
Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry | Nature Communications
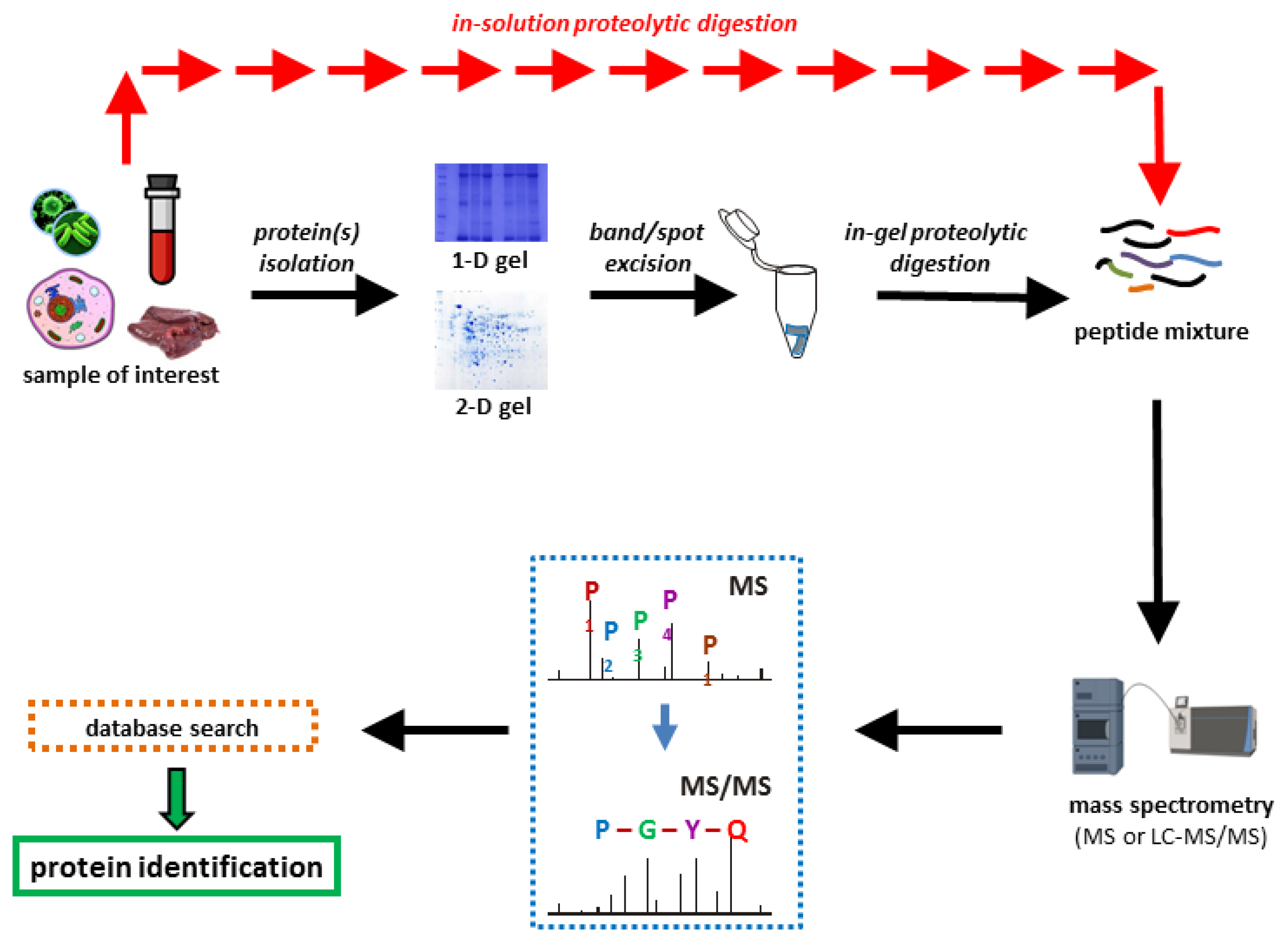
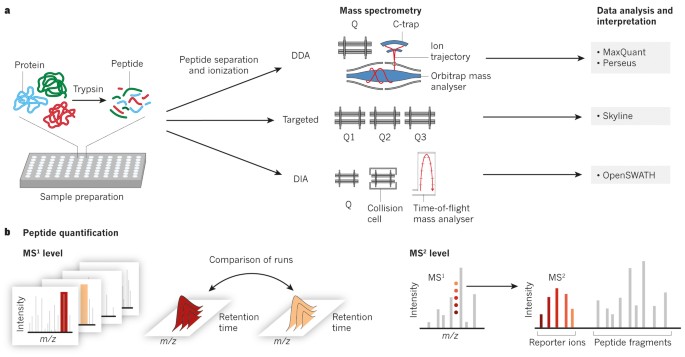
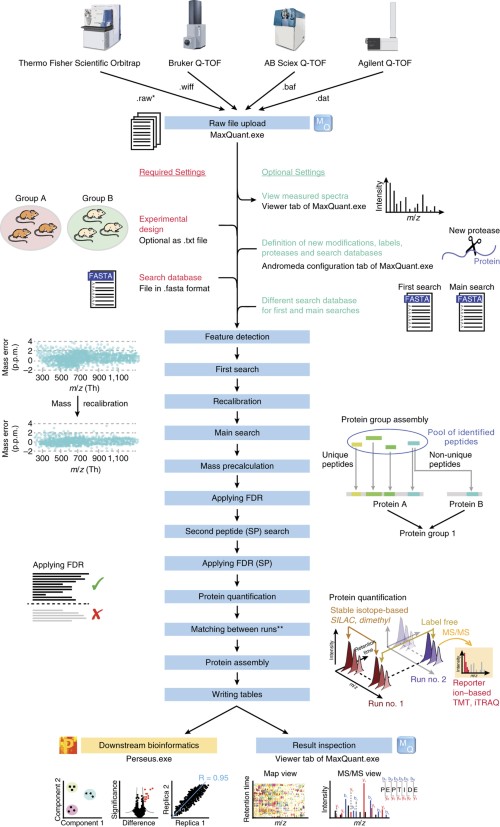
Proteomes | Free Full-Text | A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of This Field | HTML

DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput | Nature Methods
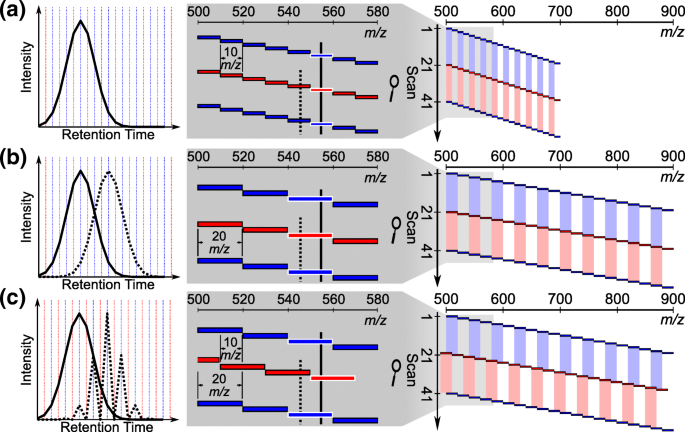
Improving Precursor Selectivity in Data-Independent Acquisition Using Overlapping Windows | SpringerLink
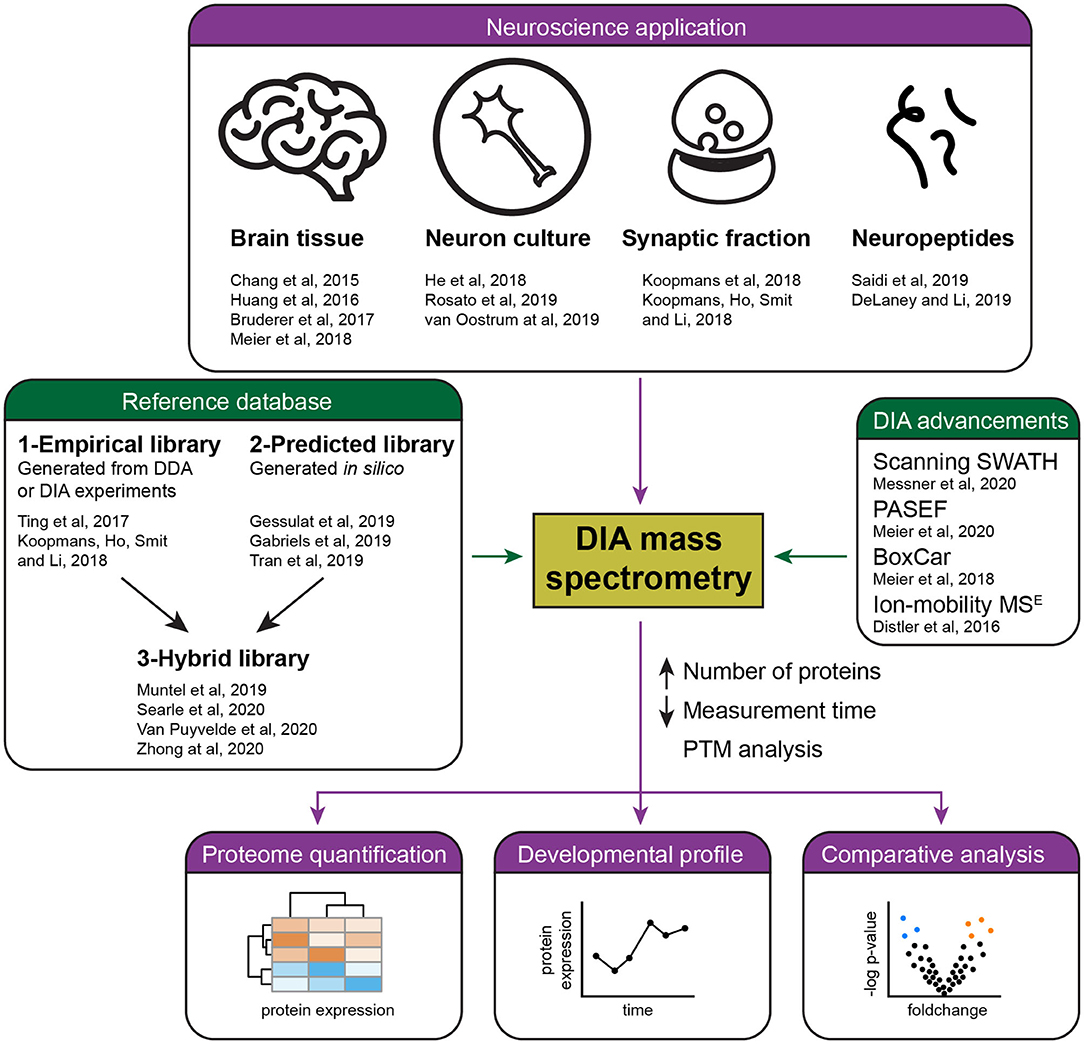
Frontiers | Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome | Molecular Neuroscience

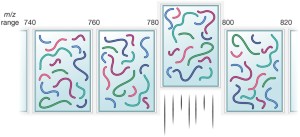
Data-independent acquisition schemes in bottom-up proteomics. (1) LC... | Download Scientific Diagram
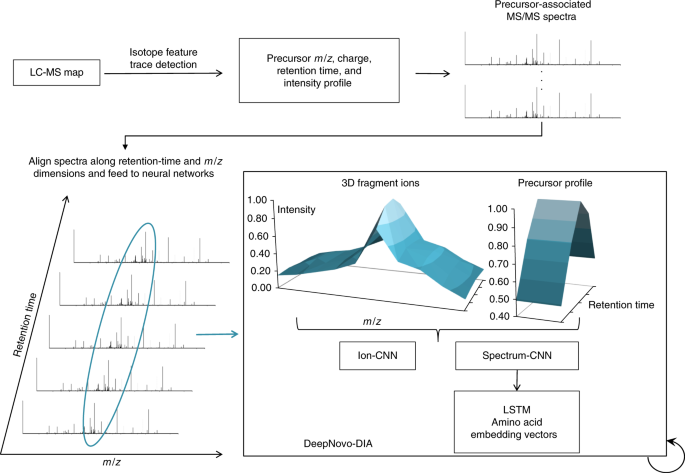
Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry | Nature Methods
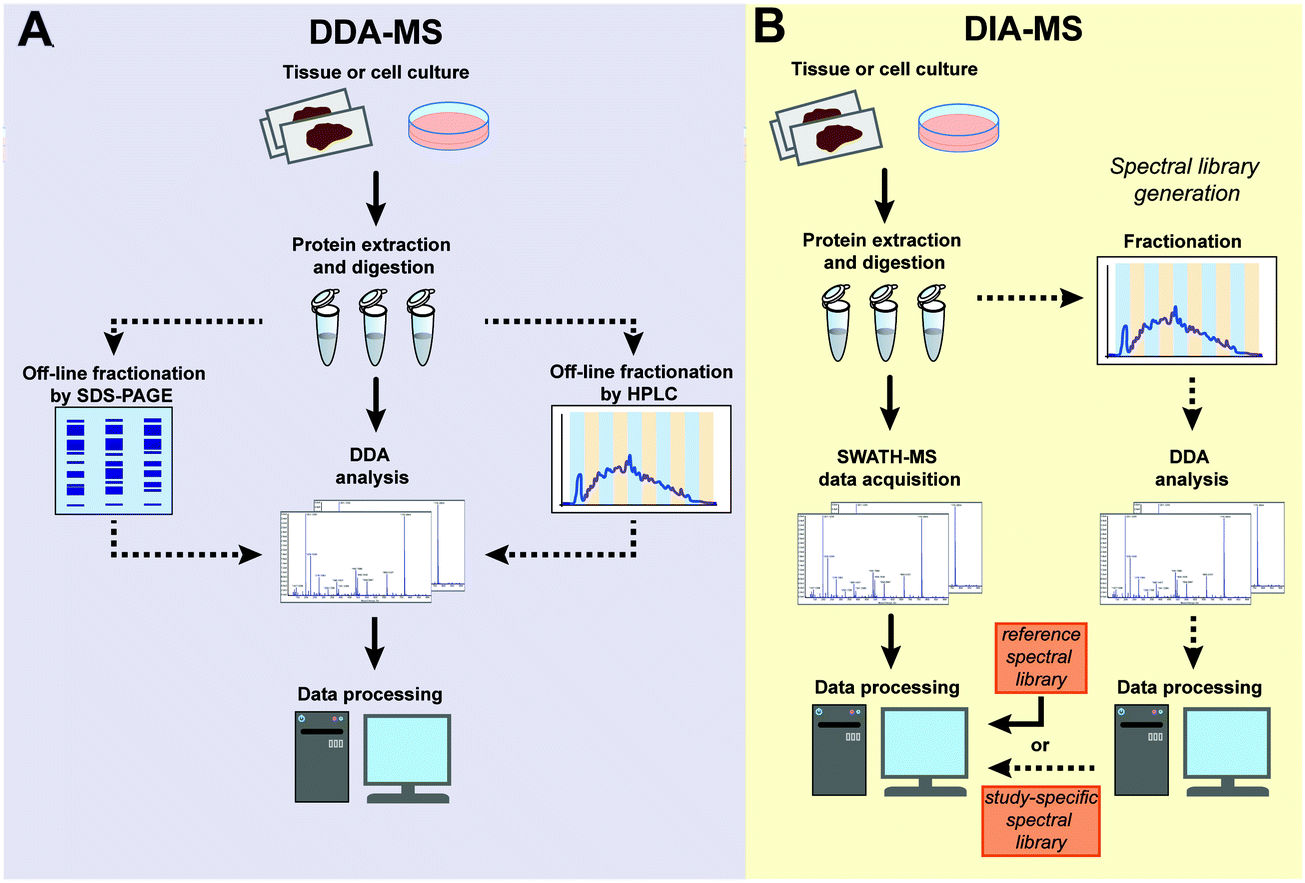
Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology - Molecular Omics (RSC Publishing) DOI:10.1039/D0MO00072H

Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology | Nature Communications
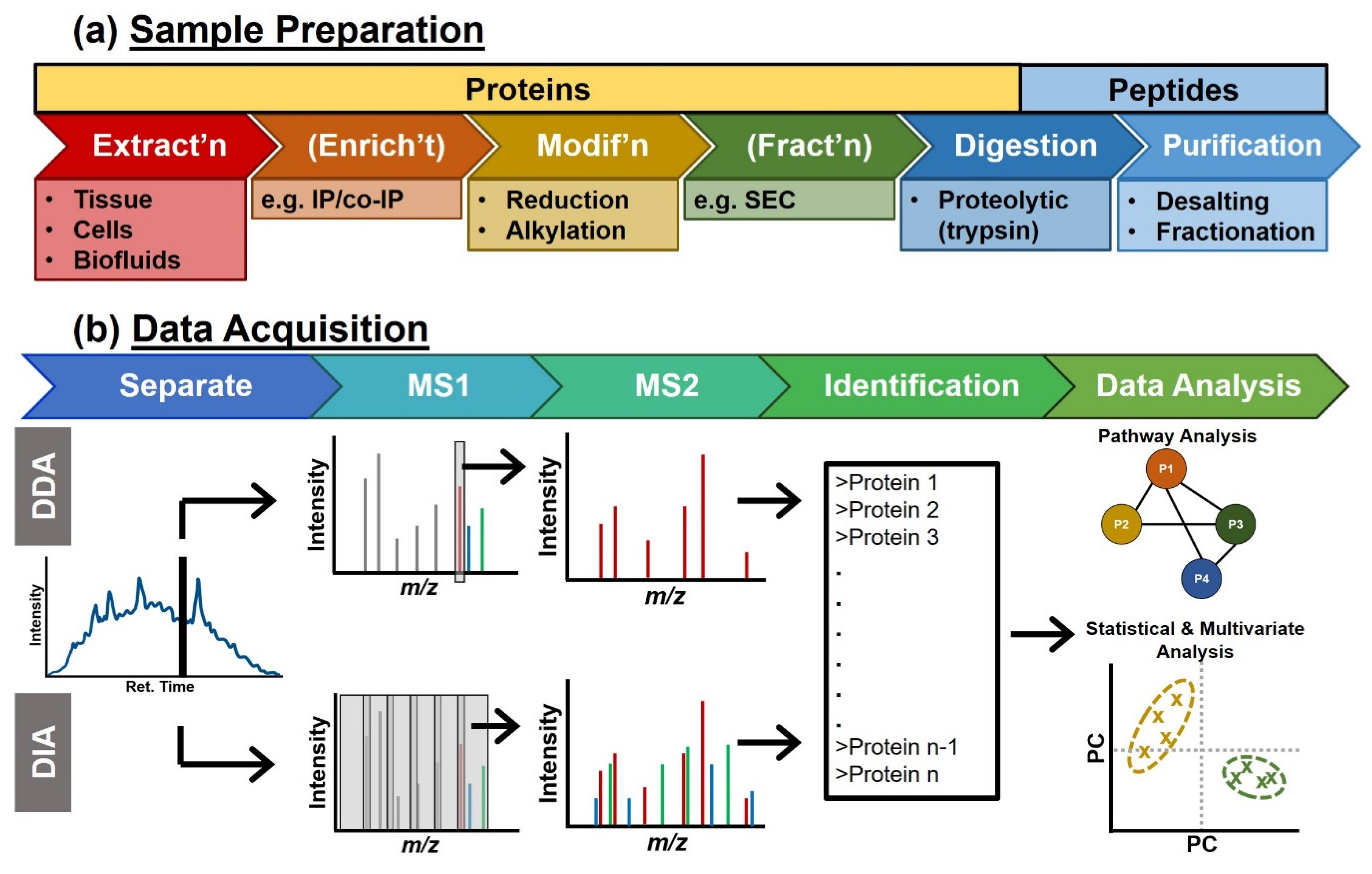
Molecules | Free Full-Text | Mass Spectrometry Advances and Perspectives for the Characterization of Emerging Adoptive Cell Therapies | HTML