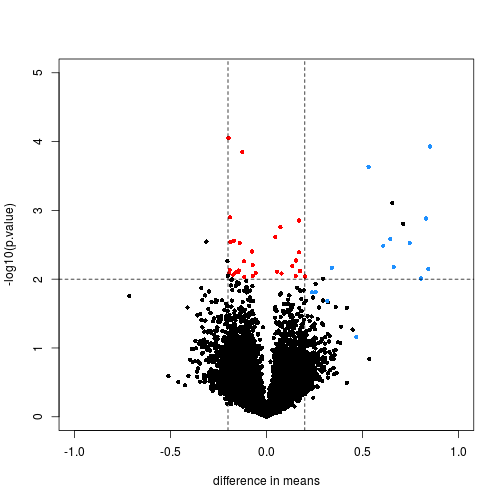
limma powers differential expression analyses for RNA-sequencing and microarray studies. - Abstract - Europe PMC

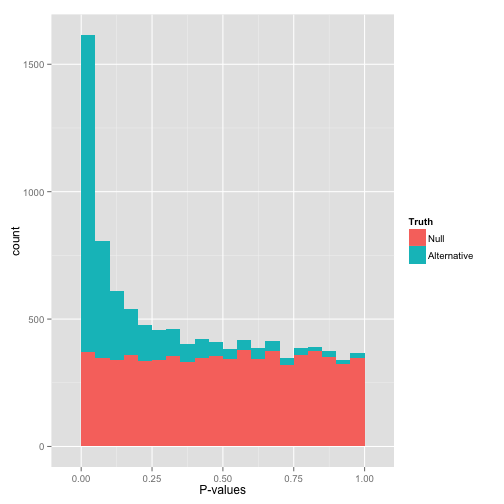
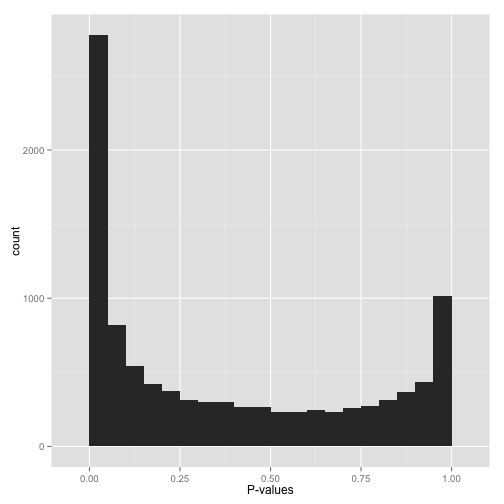
Histogram of p-values generated by conventional LIMMA and OSRR methods... | Download Scientific Diagram

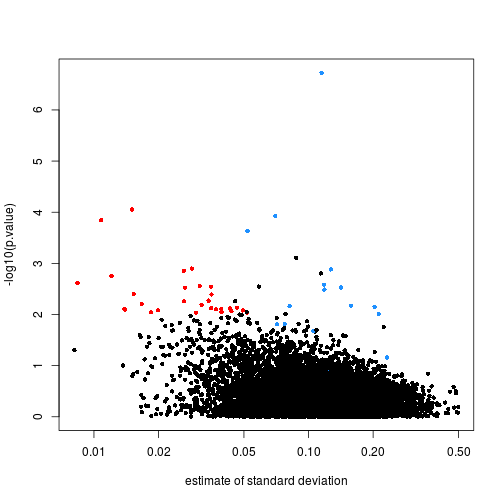
Illustration how the global p-value is calculated. On the left ((a) and... | Download Scientific Diagram

Histogram of p-values generated by conventional LIMMA and OSRR methods... | Download Scientific Diagram

QQ plots of simulated null p-values for genes in TCGA HNSC study. (A)... | Download Scientific Diagram

Histogram of p-values generated by conventional LIMMA and OSRR methods... | Download Scientific Diagram

Histogram of p-values generated by conventional LIMMA and OSRR methods... | Download Scientific Diagram

Histogram of p-values of gene expression differences from duplicate... | Download Scientific Diagram